In this case, the above likelihood function may be modified as follows:
\[\mathcal{L}(X;\theta) = \prod_{i=1}^{n} f_{\theta}(B_i-A_i)\] , where \(A^L, A^R, B\) present the times for lower end and upper bound on the potential dates of symptom onset of the infector, and the symptom onset time of the infectee, respectively.
\[f^t_{\theta}(B_i-A_i) = \frac{f_{\theta}(B_i-A_i)}{F(T-A_i)}\]
set.seed (42 )<- 1000 <- 2.2 <- 3.3 <- data.frame (A= sample (0 : 30 , size= n, replace= TRUE ))<- rgamma (n, shape= shape_true, scale= scale_true)$ B <- df$ A + simax (df$ B)
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.3967 13.9763 22.4103 22.4031 30.3811 57.0366
<- 35 <- df$ B < Tmax<- df[under_Tmax,]<- function (parms, A, B) - sum (dgamma (B- A, shape= parms[[1 ]], scale= parms[[2 ]], log= TRUE ))= optim (par= c (1 ,2 ), fn= nll, A= newdf$ A, B= newdf$ B,method = "Nelder-Mead" ,control = list (maxit= 2e4 , reltol= 1e-15 ))
$par
[1] 2.366494 2.735134
$value
[1] 2389.103
$counts
function gradient
101 NA
$convergence
[1] 0
$message
NULL
<- function (parms, A, B, Tmax) - sum (log (dgamma (B- A, shape= parms[[1 ]], scale= parms[[2 ]])/ pgamma (Tmax- A, shape= parms[[1 ]], scale= parms[[2 ]])))= optim (par= c (1 ,2 ), fn= nll_right_trunc, A= newdf$ A, B= newdf$ B,Tmax= Tmax,method = "Nelder-Mead" ,control = list (maxit= 2e4 , reltol= 1e-15 ))
$par
[1] 2.230993 3.230118
$value
[1] 2317.955
$counts
function gradient
115 NA
$convergence
[1] 0
$message
NULL
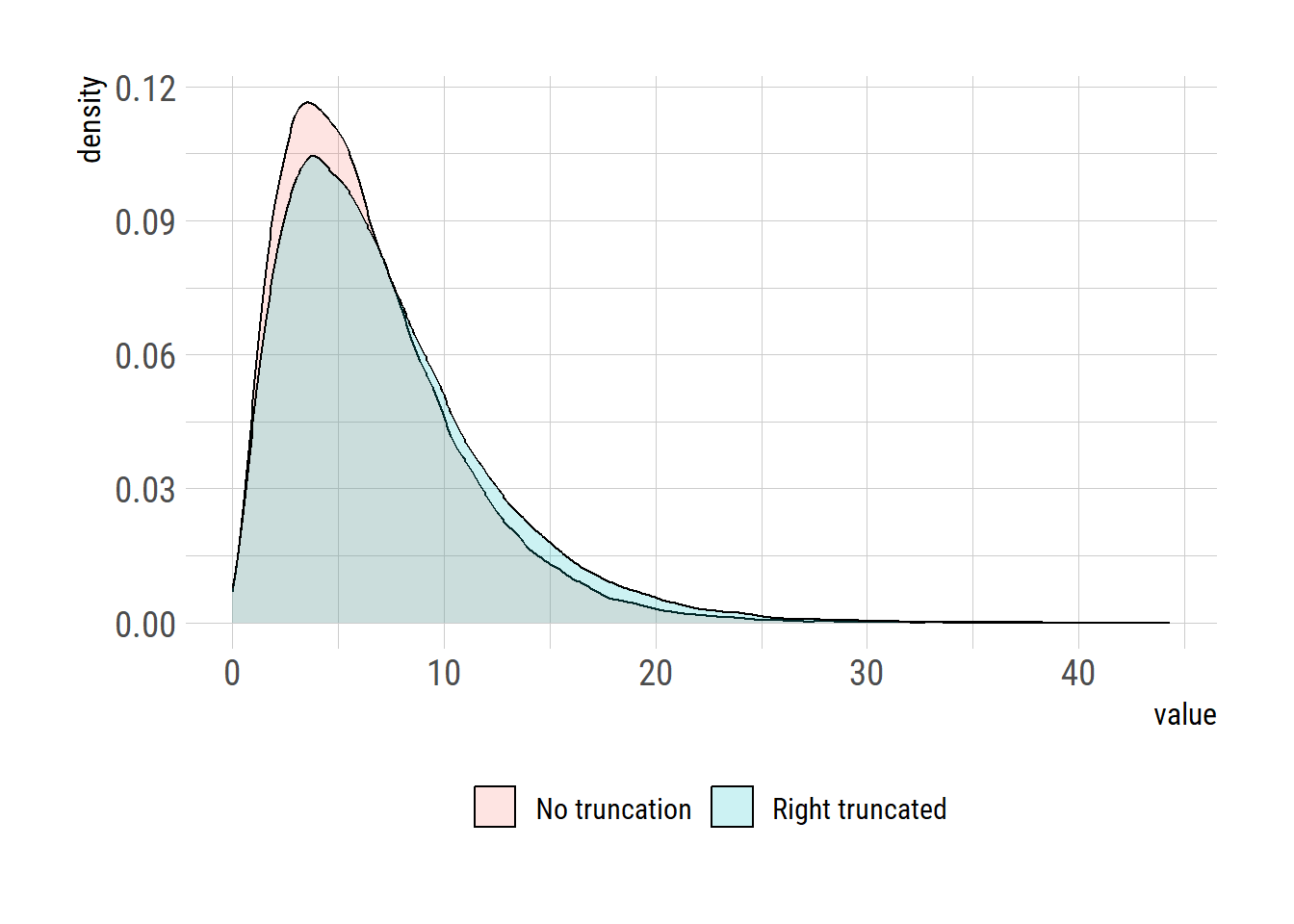
<- 1e5 <- rgamma (n, shape= res1$ par[[1 ]], scale= res1$ par[[2 ]])<- rgamma (n, shape= res2$ par[[1 ]], scale= res2$ par[[2 ]])summary (x1)
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.01066 3.36629 5.58653 6.47582 8.62659 44.32536
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.0263 3.6574 6.1697 7.2078 9.6218 43.8174
= data.frame (model= rep (c ("No truncation" , "Right truncated" ), each= n), val= c (x1,x2))library (ggplot2):: loadfonts ()theme_set (hrbrthemes:: theme_ipsum_rc (base_size= 14 , subtitle_size= 16 , axis_title_size= 12 ))|> ggplot (aes (x= val, fill= model))+ geom_density (alpha= 0.2 ) + labs (x= "value" , y= "density" )+ theme (legend.position = "bottom" , legend.title = element_blank ())# ggsave("right_trunc.png", gg, units="in", width=3.4*2, height=2.7*2)