Data
I will use the cars data with give the speed of cars and the distances taken to stop.
d <- datasets::cars
m <- lm(dist ~ speed, data=d)
summary(m)
Call:
lm(formula = dist ~ speed, data = d)
Residuals:
Min 1Q Median 3Q Max
-29.069 -9.525 -2.272 9.215 43.201
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -17.5791 6.7584 -2.601 0.0123 *
speed 3.9324 0.4155 9.464 1.49e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 15.38 on 48 degrees of freedom
Multiple R-squared: 0.6511, Adjusted R-squared: 0.6438
F-statistic: 89.57 on 1 and 48 DF, p-value: 1.49e-12
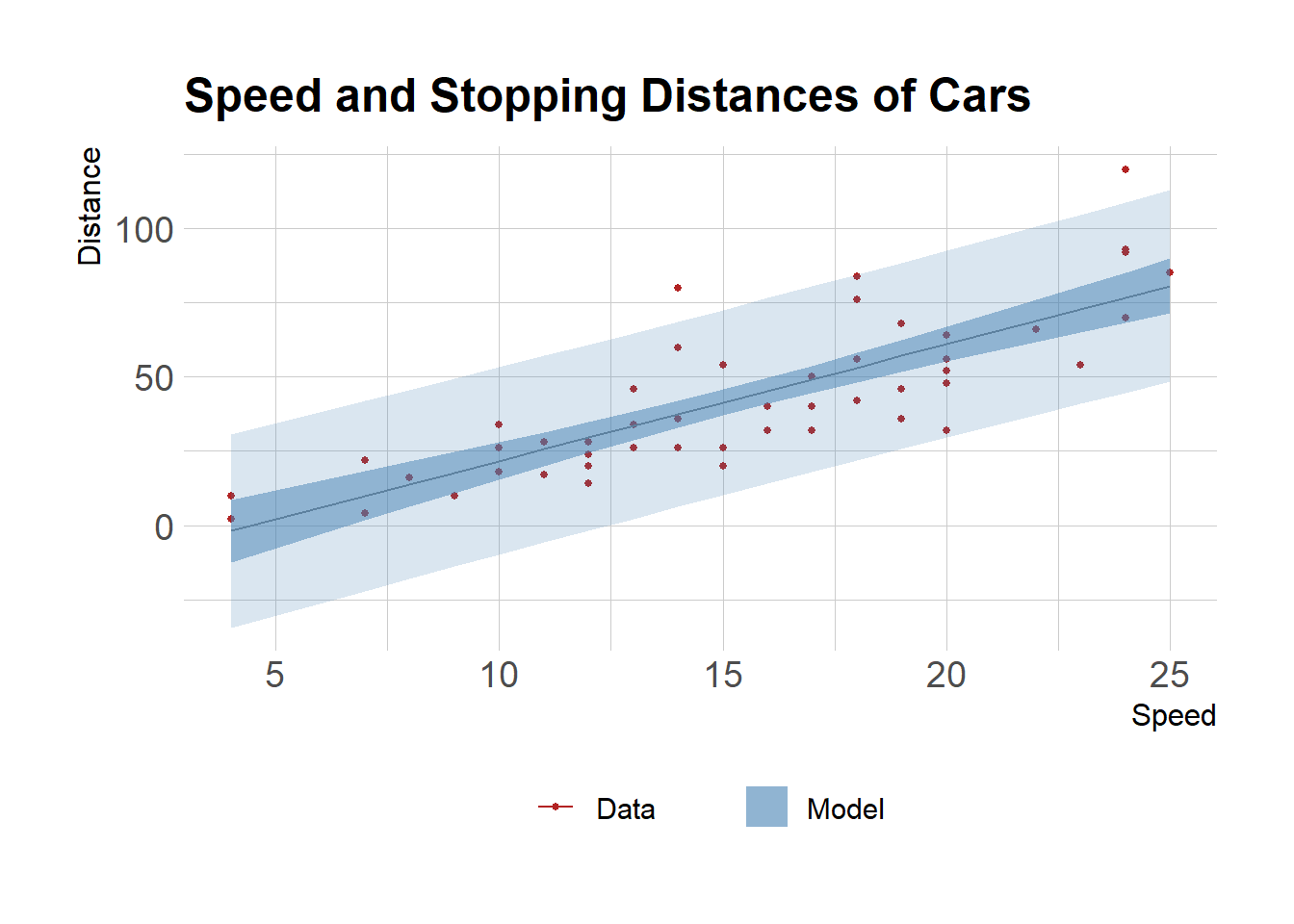
Plot
Plot estimates with confidence and prediction intervals
pred <- predict(m, interval="prediction", level=0.95) # prediction interval
conf <- predict(m, interval="confidence", level=0.95) # confidence interval
mdat <- m$model
mdat$pred_estimate <- pred[,1]
mdat$pred_lb <- pred[,2]
mdat$pred_ub <- pred[,3]
mdat$conf_estimate <- conf[,1]
mdat$conf_lb <- conf[,2]
mdat$conf_ub <- conf[,3]
mdat$residuals <- residuals(m)
library(ggplot2)
library(dplyr)
theme_set(hrbrthemes::theme_ipsum_rc(base_size=14, subtitle_size=16, axis_title_size=12))
pltcar <- mdat %>%
ggplot(aes(speed, dist))+
# geom_point(aes(size = abs(m$residuals)))+
geom_point(aes(color="Data"), size = 1)+
geom_line(aes(y=pred_estimate, color="Model"))+
geom_ribbon(aes(ymax=pred_ub, ymin=pred_lb, fill="Model"), alpha=0.2)+
# geom_line(aes(y=conf_estimate), color="steelblue")+
geom_ribbon(aes(ymax=conf_ub, ymin=conf_lb, fill="Model"), alpha=0.5)+
scale_color_manual("", values=c("Data"="firebrick"))+
scale_fill_manual("", values=c("Model"="steelblue"))+
labs(x="Speed", y="Distance", title="Speed and Stopping Distances of Cars")+
theme(legend.position="bottom")
# ggsave("plot_car.png", pltcar)
pltcar
optim function
Now let’s take an alternative approach to write down the likelihood function and maximize it using the optim function
# define our likelihood function we like to optimize
negloglik <- function(par, y, X){
sigma <- par[1]
beta <- par[-length(par)]
mu <- X %*% beta
- sum(dnorm(y, mean=mu, sd=sigma, log=TRUE), na.rm=T)
}
X = model.matrix(m)
init = c(coef(m), sigma=summary(m)$sigma)
# check
# negloglik(par=init, y=d$y, X=X)
fit <- optim(par=init,
fn=negloglik,
y=d$dist,
X=X,
control=list(reltol=1e-6))
Let’s compare the results.
(Intercept) speed sigma
-17.579095 3.932409 15.379587
(Intercept) speed
-17.579095 3.932409
