reulermultinom2 <- function (m=2, size, rate, dt) {
trans <- matrix(NA, nrow=m, ncol=length(rate))
p <- 0.0 # total event rate
if ((size < 0.0) | (dt < 0.0) | (floor(size+0.5) != size)) {
for (k in seq_along(rate)) {
trans[k] = NaN
}
return(trans)
}
if (sum(rate < 0.0) > 0){
stop("Negative rates are not allowed")
}
else {
p <- sum(rate)
}
if (p > 0.0) {
for (i in 1:m) {
tmpsize <- rbinom(1, size = size, prob = (1-exp(-p*dt))) # total number of events
tmpp <- p
for (k in 1:(length(rate)-1)) {
trans[i, k] = rbinom(1, tmpsize, rate[k]/tmpp)
tmpsize = tmpsize - trans[i, k];
tmpp = tmpp - rate[k];
}
trans[i, length(rate)] = tmpsize;
}
}
return(trans)
}Multinomial distribution
A simple particle filter in R
When implementing a model of stochastic disease transmission, one has to deal with a situation in which multiple events are possible. For example, susceptible people may become infected, remain susceptible, or die from other causes. In R, one could use rmultinorm as long as one can assign a probability for each event. Here, however, we implement a function from scratch. One way is to follow the approach of Aaron King, author of the pomp package. His method is implemented in C and I adapted it to R and C++ while removing many of its auxiliary functions (e.g., checking the validity of the inputs).
Let’s compare it with the original function provided in the pomp package
x <- t(pomp::reulermultinom(1e5, 100, rate=c(1,2), dt=0.05))
y <- reulermultinom2(1e5, 100, rate=c(1,2), dt=0.05)
xy <- as.data.frame(cbind(x, y))
names(xy) <- c("pomp_var1", "pomp_var2", "var1", "var2")
apply(xy, 2, summary) pomp_var1 pomp_var2 var1 var2
Min. 0.00000 0.00000 0.0000 0.00000
1st Qu. 3.00000 7.00000 3.0000 7.00000
Median 4.00000 9.00000 4.0000 9.00000
Mean 4.64097 9.27939 4.6419 9.28399
3rd Qu. 6.00000 11.00000 6.0000 11.00000
Max. 16.00000 25.00000 15.0000 24.00000The speed difference is quite substantial.
library(microbenchmark)
microbenchmark(pomp::reulermultinom(100, 100, rate=c(1,2), dt=0.05), reulermultinom2(100, 100, rate=c(1,2), dt=0.05))Unit: microseconds
expr min lq
pomp::reulermultinom(100, 100, rate = c(1, 2), dt = 0.05) 39.702 41.4505
reulermultinom2(100, 100, rate = c(1, 2), dt = 0.05) 377.301 384.9005
mean median uq max neval cld
48.18708 43.651 48.0015 92.801 100 a
561.12002 401.851 641.1015 6275.101 100 bRewrite the function in C++ using Rcpp.
Rcpp::cppFunction("NumericMatrix reulermultinom_cpp(int m, double size, NumericVector rate, double dt) {
int ncol = rate.size();
NumericMatrix trans(m, ncol);
double p = sum(rate); //total event rate
for (int i = 0; i < m; i++) {
double tmpp = p;
double tmpsize = R::rbinom(size, (1-exp(-tmpp*dt))); // total number of events
for (int k = 0; k < (ncol-1); k++) {
double tr = R::rbinom(tmpsize, rate(k)/tmpp);
trans(i, k) = tr;
tmpsize = tmpsize - trans(i, k);
tmpp = tmpp - rate(k);
}
trans(i, (ncol-1)) = tmpsize;
}
return(trans);
}")library(microbenchmark)
microbenchmark(pomp::reulermultinom(1e5, 100, rate=c(1,2), dt=0.05), reulermultinom_cpp(1e5, 100, rate=c(1,2), dt=0.05))Unit: milliseconds
expr min lq
pomp::reulermultinom(1e+05, 100, rate = c(1, 2), dt = 0.05) 24.9228 27.61285
reulermultinom_cpp(1e+05, 100, rate = c(1, 2), dt = 0.05) 23.9839 25.87615
mean median uq max neval cld
51.27772 41.71065 66.0408 131.8367 100 a
51.02618 43.38275 67.6930 120.4940 100 ax <- t(pomp::reulermultinom(1e5, 100, rate=c(1,2), dt=0.05))
y <- reulermultinom_cpp(1e5, 100, rate=c(1,2), dt=0.05)
xy <- as.data.frame(cbind(x, y))
names(xy) <- c("pomp_var1", "pomp_var2", "var1", "var2")
apply(xy, 2, summary) pomp_var1 pomp_var2 var1 var2
Min. 0.0000 0.00000 0.00000 0.00000
1st Qu. 3.0000 7.00000 3.00000 7.00000
Median 4.0000 9.00000 4.00000 9.00000
Mean 4.6498 9.28149 4.64416 9.28147
3rd Qu. 6.0000 11.00000 6.00000 11.00000
Max. 17.0000 22.00000 15.00000 24.00000library(tidyr)
xy |> pivot_longer(cols=1:4, names_to="var") -> xylong
library(ggplot2)
theme_set(hrbrthemes::theme_ipsum_rc(base_size=14, subtitle_size=16, axis_title_size=12))
extrafont::loadfonts()
ggplot(xylong)+
geom_violin(aes(x=var, y=value))+
facet_wrap(~var, nrow=1, scales="free_x")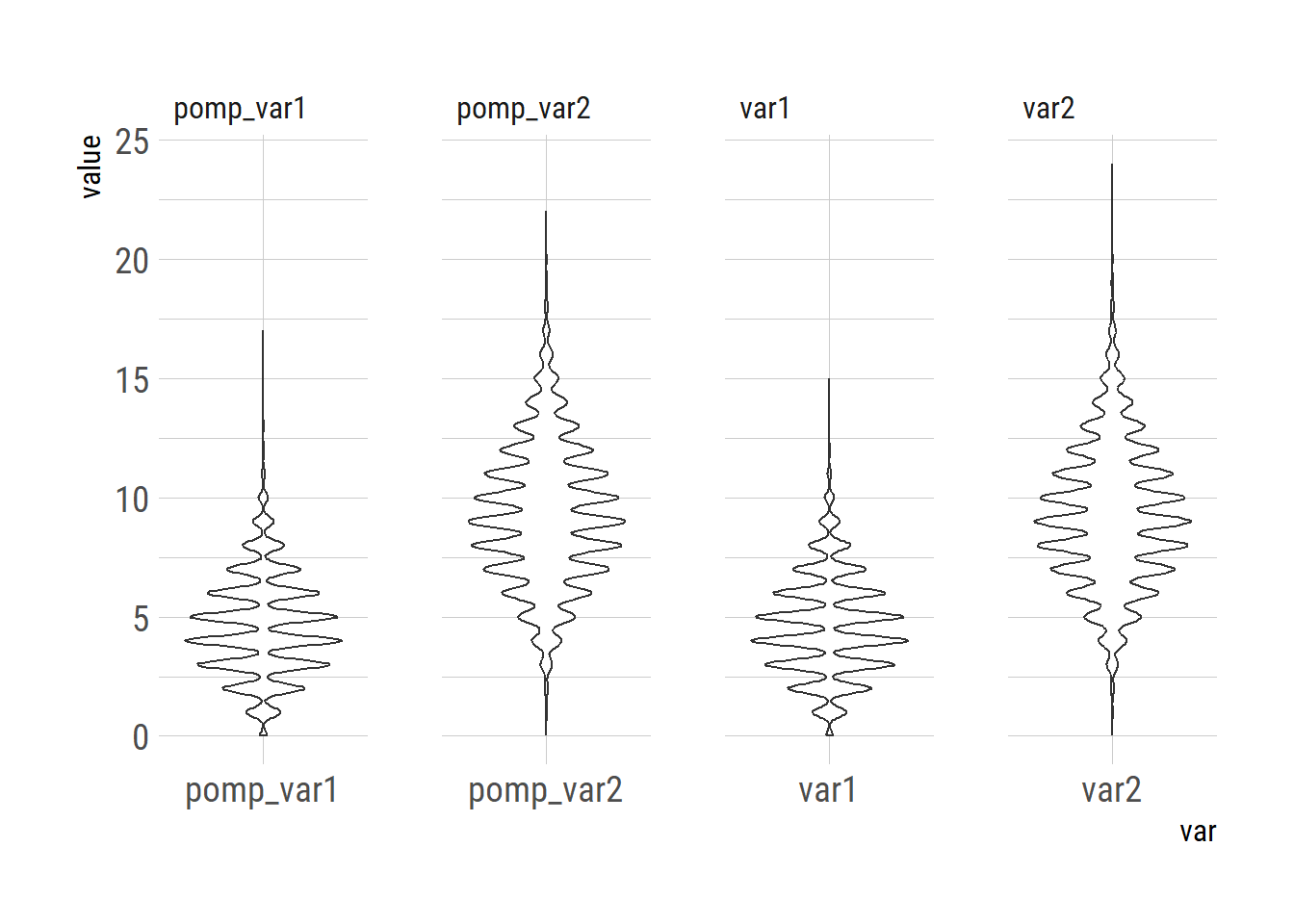
# ggsave("multinomial.png", gg, units="in", width=3.4*2, height=2.7*2)