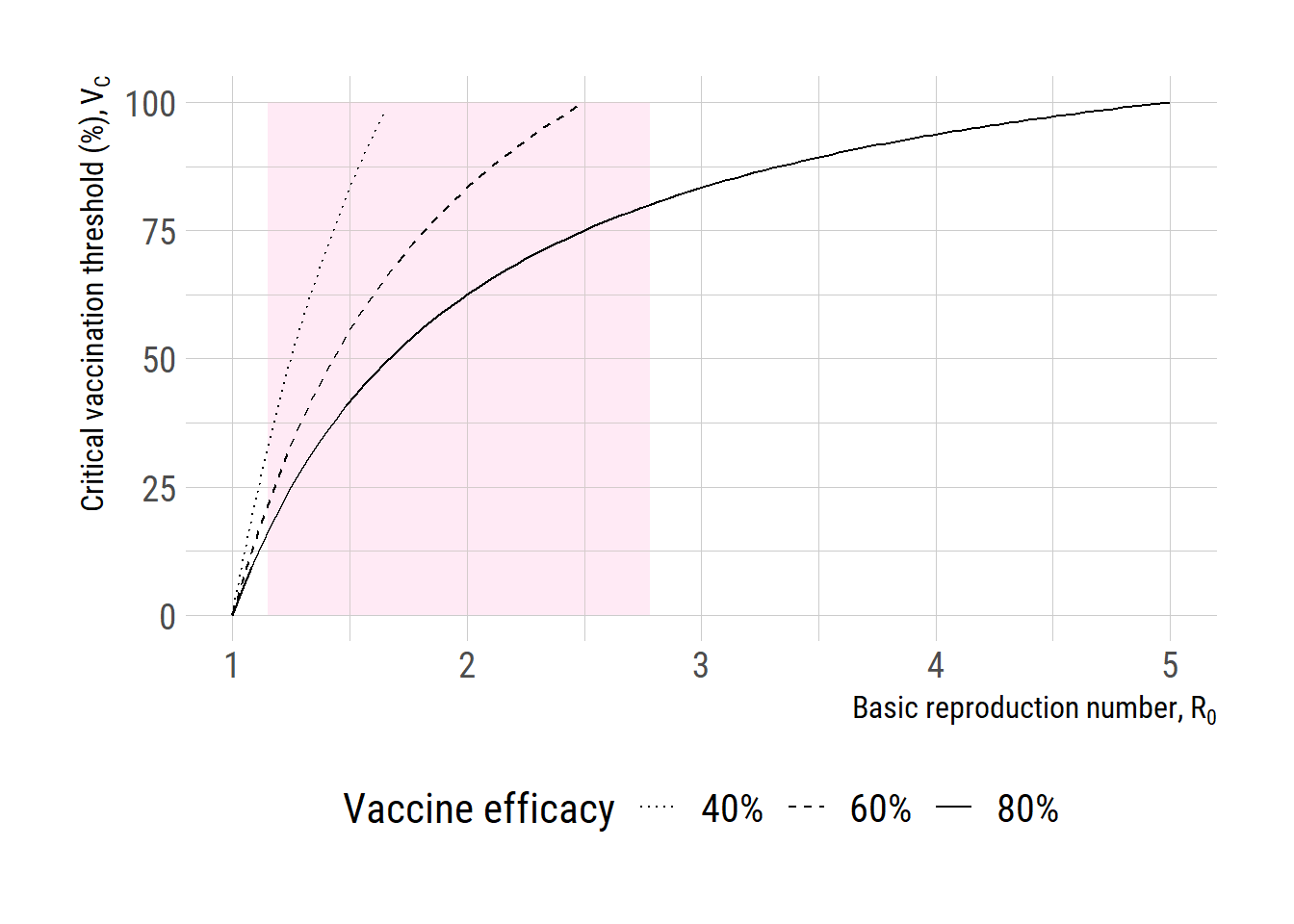
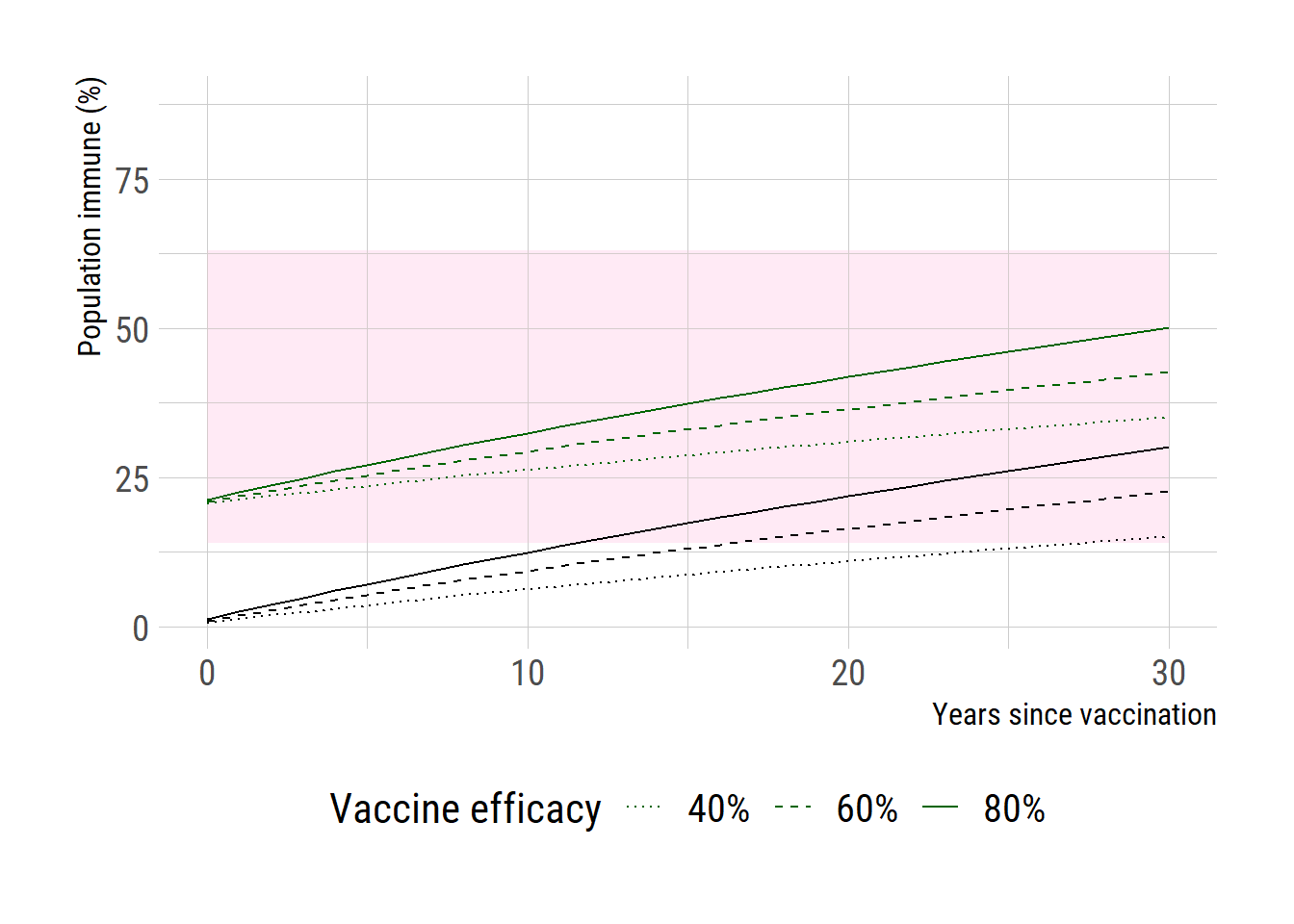library (data.table)library (readxl)library (dplyr)# d <- read_xlsx("C:/Users/jonghoon.kim/Documents/myblog/posts/critical_vacc_threshold/WPP2022_POP_F01_1_POPULATION_SINGLE_AGE_BOTH_SEXES.xlsx", sheet= "Medium variant") # d2 <- d[-(1:11),] # india <- d2[(d2$...3 == "India" & d2$...11 > 2023),] # names(india) <- c(1:2, "country", 4:10 ,"year", paste0("age_", 0:100)) # saveRDS(india, "C:/Users/jonghoon.kim/Documents/myblog/posts/critical_vacc_threshold/WPP2022_India.rds") <- readRDS ("WPP2022_India.rds" )$ year <- as.numeric (india$ year)# india[,paste0("age_", 0:100)] <- as.numeric(india[,paste0("age_", 0:100)]) <- india %>% mutate_at (paste0 ("age_" , 0 : 100 ), as.numeric)<- 2024 : 2100 # simulate <- c (0.4 , 0.6 , 0.8 )<- rep (NA , length (yrs))for (i in seq_along (yrs)) {<- sum (india[india$ year == yrs[i], paste0 ("age_" , 0 : (i-1 ))])<- sum (india[india$ year == yrs[i], paste0 ("age_" , 0 : 100 )])<- numerator / denominator<- 20 <- lapply (VE, function (z) 100 * z* prop_immune)# add the existing immunity, assumed to be 15% <- lapply (VE, function (z) 100 * z* prop_immune + existing_immune)<- data.frame (years= yrs-2024 , pi1= pop_immune_ve[[1 ]], pi2= pop_immune_ve[[2 ]], pi3= pop_immune_ve[[3 ]], pi4= pop_immune_ve_added[[1 ]], pi5= pop_immune_ve_added[[2 ]], pi6= pop_immune_ve_added[[3 ]]):: loadfonts ("win" , quiet= TRUE )theme_set (hrbrthemes:: theme_ipsum_rc (base_size= 14 , subtitle_size= 16 , axis_title_size= 12 ))<- ggplot (df, aes (x = years)) + geom_rect (aes (xmin= 0 , xmax= 30 , ymin= 14 , ymax= 63 ),fill = "pink" , alpha= 0.01 )+ geom_line (aes (y= pi3, linetype= "80%" )) + geom_line (aes (y= pi2, linetype= "60%" )) + geom_line (aes (y= pi1, linetype= "40%" )) + geom_line (aes (y= pi6, linetype= "80%" ), color= "dark green" ) + geom_line (aes (y= pi5, linetype= "60%" ), color= "dark green" ) + geom_line (aes (y= pi4, linetype= "40%" ), color= "dark green" ) + scale_x_continuous (limits= c (0 ,30 ))+ scale_linetype_manual ("Vaccine efficacy" , values= c ("80%" = "solid" ,"60%" = "dashed" , "40%" = "dotted" ))+ # labs(y="Critical vaccination threshold", x=expression(R[0])) + labs (y= "Population immune (%)" , x= "Years since vaccination" ) + theme (text = element_text (size= 16 ),axis.text = element_text (size= 16 ),legend.text= element_text (size= 15 ), legend.position = "bottom" )